Segmentation of cellular compartments
Segmentation adapted to each organelle
Every cellular compartment has a specific morphology, a fibrous structure cannot be thus segmented in the same way as a spot (like a kinetocore) or a round structure (like a nucleus). The detection of a compartment is a complex process which requires to parametrize finely segmentation tools to analyse correctly the fluorescent signal. Generally speaking, these groups of components are segmented in the same way (list is not exhaustive):
- Fibers: strands of actine, microtubules, intermediate strands, extracellular matrix
- Complex compartments: Golgi apparatus, endoplasmic reticulum, chromosomes during the mitosis,mitotic spindle, mitocondria, ribosome
- Spots: endosomes, lysosomes, MOC, vacuoles, vesicles
- Membrane: cytoplasmic membrane, nuclear membrane
Examples of segmentations adapted to centrosome, Golgi and nuclei
The segmentation of compartments allows to analyze their morphology (size, shape, intensity, position in the cell) and their position compared with other compartments.
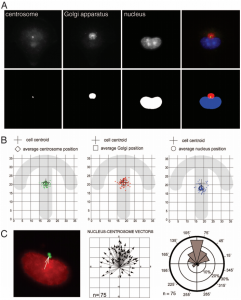
The following figure illustrates the position of compartments in cells plated into micropatterns. Three cellular compartments are separately segmented: centrosomes, Golgi apparatus and nuclei. The figure A shows every compartment and its segmentation. The figure B shows the distributions of compartments in the space (centrosome in green, Golgi in red and nuclei in blue). The figure C represents vectors nuclei – centrosomes and their directions
References
Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity.Théry M, Racine V, Piel M, Pépin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M. Proc Natl Acad Sci U S A, 103(52):19771-6, 2006.
Interested in softwares or services?
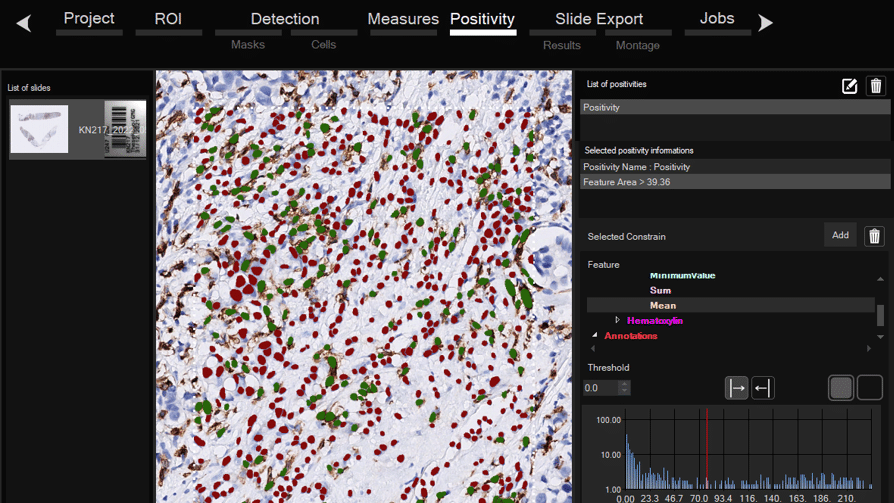
Software for histological samples and TMA
Automatically detects and quantifies your histological slides easily, without requiring bioinformatics expertise. IHC/Fluorescence/Deep learning
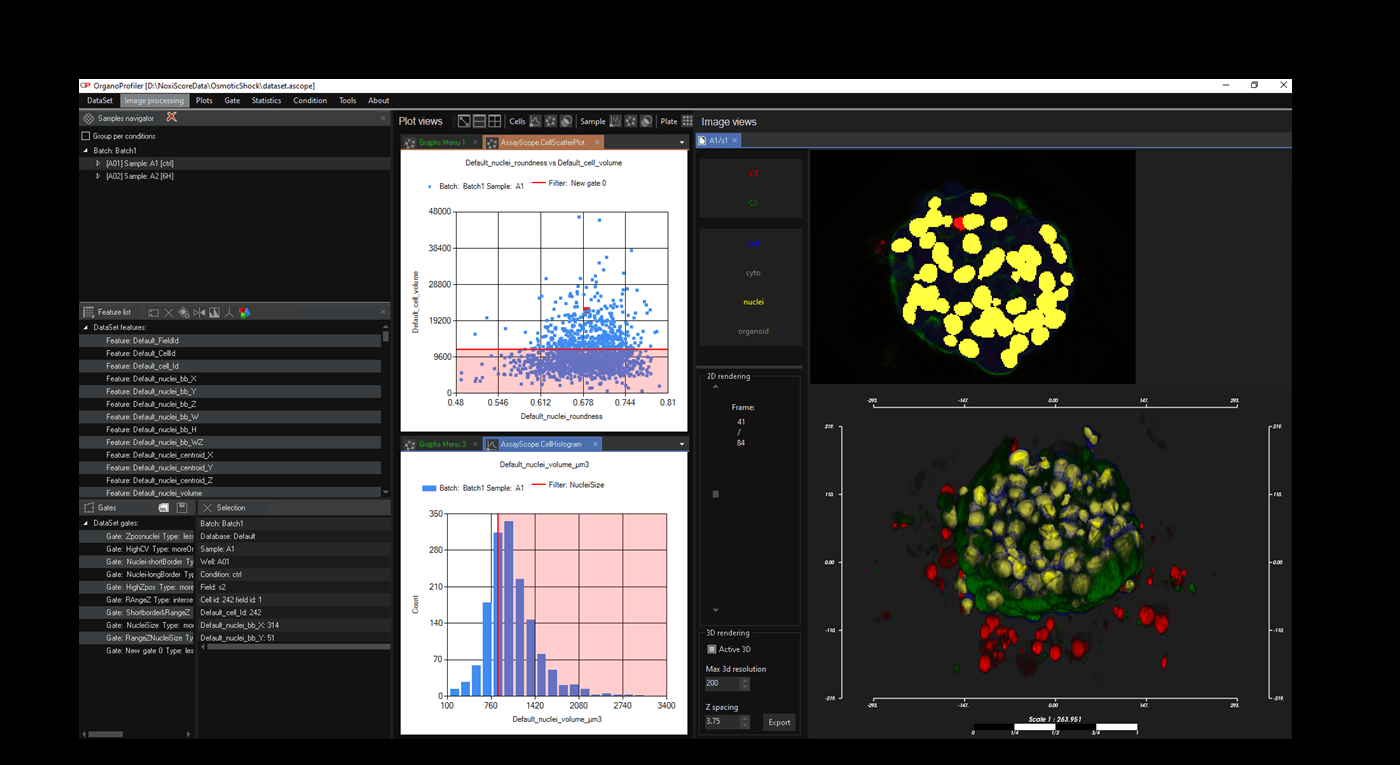
Software for organoid, spheroid and 3D cell culture and tissue
Automatic 3D segmentation and high-content screening analysis software to navigate your assays and monitor drug effects
Services of analysis and characterization
- Image analysis in patients studies
- Characterization of drug/compounds effects
- Automatic detection of rare events

Software development for biomedical imaging
- Customized software solutions with ergonomics UIX
- AI for automatic analysis
- Industrialization of analyses in laboratories
QuantaCell, Campus Balard, IHU
Contact
+33 (0) 9 83 33 81 90
300 av. du Pr Emile Jeanbrau
34090 Montpellier, France