Merging heterogeneous information
Merging microarray data with public bio-informatic information
The mass of information on the biological and chemical data accessible via the Internet is huge. This information can be used to enrich the experimental results.
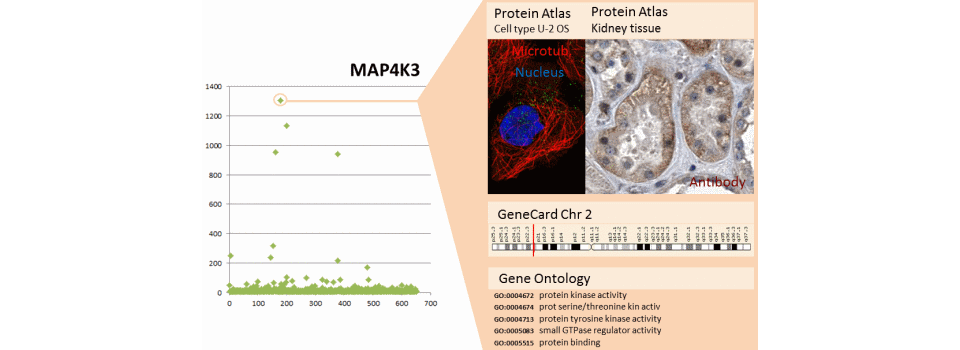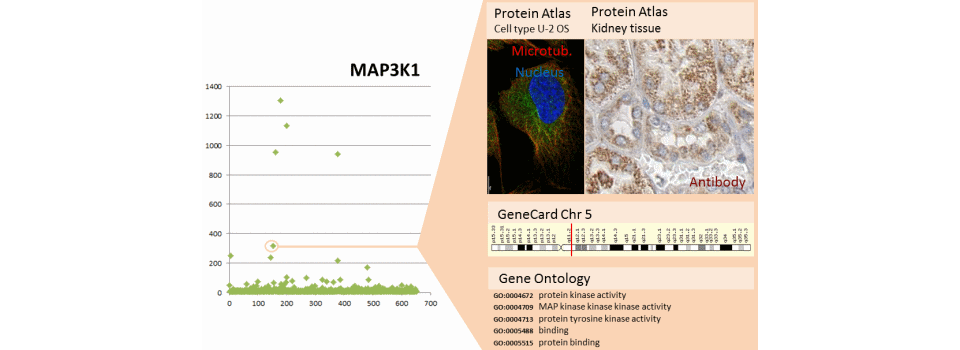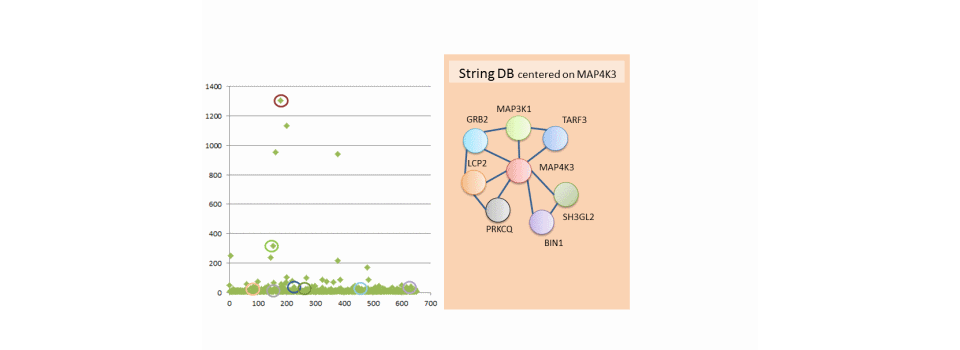
The example shows the results of a microarray kinome complemented by public data. Microarray data are presented on the left. The user selects a gene (MAP4K3 and MAP3K1) and public data appear in the right window to cross the user information with public information.
Access to biomedical information in public databases
Public information of the selected genes are automatically retrieved from several public knowledge bases:
- GeneCard (http://www.genecards.org/)
- Gene Ontology (accessed via GeneCard)
- HumanProtein Atlas (http://www.proteinatlas.org/) provides the localisation of proteins in cellular models, in tissues of many cancerous or normal organs
- EMAGE (http://www.emouseatlas.org/) provides protein expression in mouse embryos
- STRING (http://string-db.org/) provides interactome data
Similarly, the interactome MAP4K3 is displayed in the right window and each gene in the interactome is selected from the microarray data.
This information is integrated into the GUI of the software for optimal interactivity.
This strategy allows the user to have a maximum of elements and better interpretation of the experimental results
Interested in softwares or services?
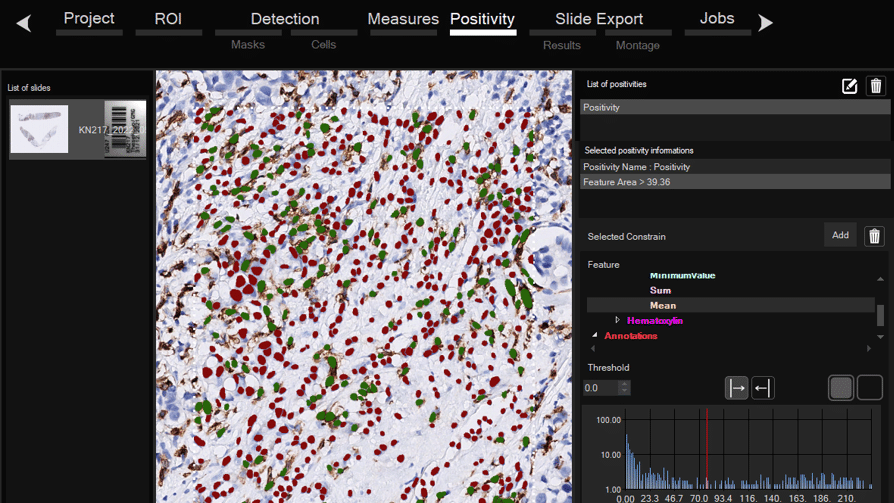
Software for histological samples and TMA
Automatically detects and quantifies your histological slides easily, without requiring bioinformatics expertise. IHC/Fluorescence/Deep learning
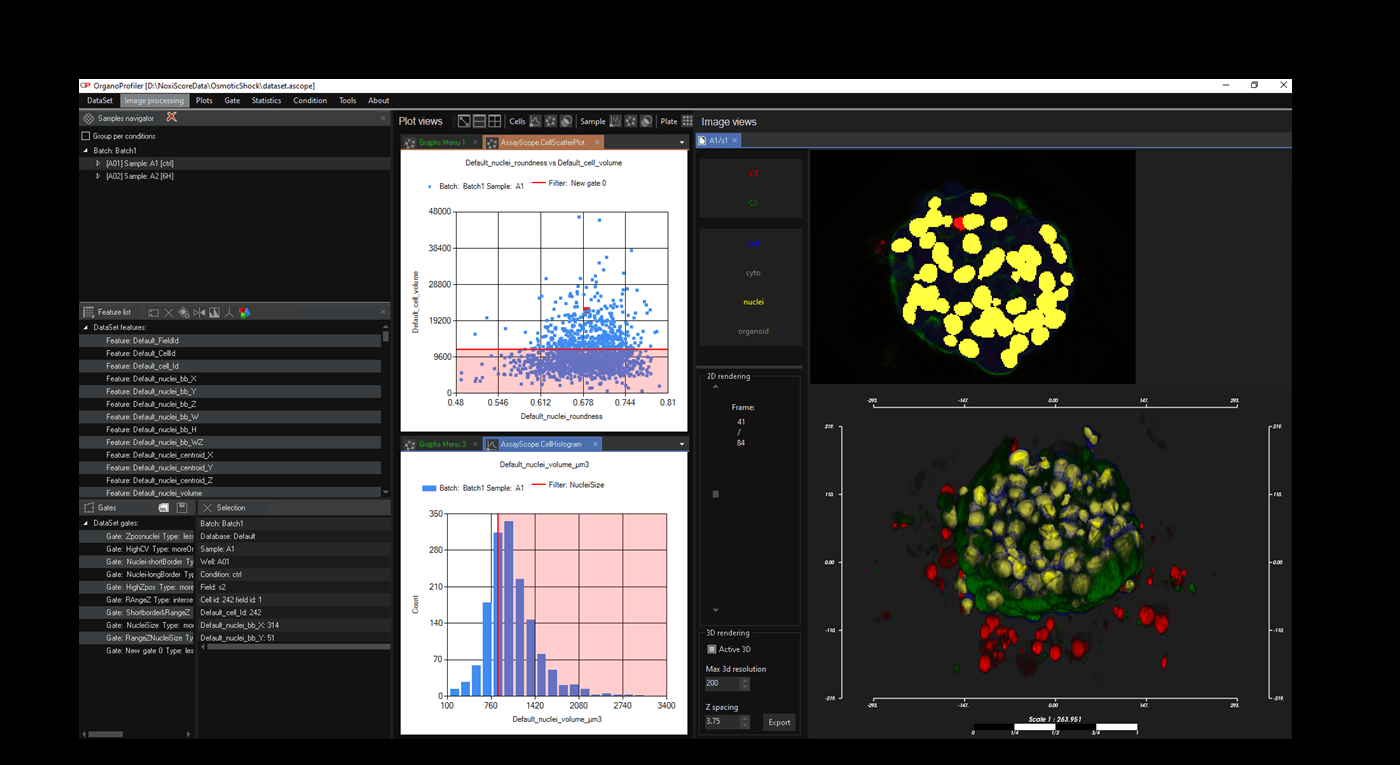
Software for organoid, spheroid and 3D cell culture and tissue
Automatic 3D segmentation and high-content screening analysis software to navigate your assays and monitor drug effects
Services of analysis and characterization
- Image analysis in patients studies
- Characterization of drug/compounds effects
- Automatic detection of rare events

Software development for biomedical imaging
- Customized software solutions with ergonomics UIX
- AI for automatic analysis
- Industrialization of analyses in laboratories
Registered address
73 Allée Kléber,
34000 Montpellier, France
Laboratory (in Montpellier’s IHU)
300 av. du Pr Emile Jeanbrau 34090 Montpellier, France
Contact
+33 (0) 9 83 33 81 90