Tissue analysis
Quantitative histology and tissue analysis
The automatic quantification of tissues serves to reproduce the interpretation of pathologists, by means of algorithms. The tissue sections are arranged on microscope slides and digitized with a slide scanner. Automatic quantification is a complex task because:
- Images are huge: slide scanner generates virtual slides combining thousands of microscope fields (tiles) that correspond to the entire tissue sample
- Slide to slide variability: the inclusion steps, staining steps and acquisitions steps can lead to important differences between identical slides. Thus it is important to process carefully images in order to reduces slide to slide differences. Internal normalization can help to standardize colors and intensities
- Several segmentation steps are required to achieve good quantification: sample segmentation (to remove the background of the sample), tissue structure segmentation (like epithelium area, necrotic area, lumens, tissue lesions), cells segmentation (to obtain individual intensity, shape, number of spots)
- Result visualization is not trivial: the visual inspection of slide quantification is very important and the user has to verify the results at different resolutions. He has to be able to navigate with ergonomic tools to zoom in, zoom out and pane the original image and superimpose resulting segmentation
- In many situation, the analysis has to be guided by the expert or the anatomopathologist. They define region of interest, include or exclude areas according to their understanding. So it is important to combine automatic quantification with expert supervision
The tissue slides and Tissue Micro Array (TMA)
A tissue slide can contain :

Histological stainings
Common stainings are :
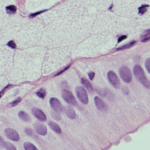
Histochemistry: staining based on chemical reactions between reagents and components of the tissue under study (e.g.: hematoxylin-eosin staining, Periodic Acid-Shiff (PAS), Masson’s Trichrome de Masson)
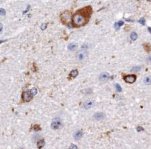
Immunohistochemistry (IHC): Immunological recognition of any given antigenic target (e.g. protein). Antibodies are coupled to an enzyme that convert a chromogenic substrate in a colored precipitate. Detection can be direct (enzyme-coupled primary antibody to antigen) or indirect (enzyme-coupled secondary antibody recognizes non-coupled primary antibody).
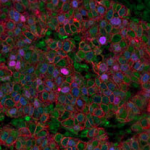
Immunofluorescence (IF): Immunological recognition of any given antigenic target (e.g. protein). Antibodies are coupled to a fluorochrome. Detection can be direct (fluorochrome-coupled primary antibody to antigen) or indirect (fluorochrome-coupled secondary antibody recognizes non-coupled primary antibody)
In situ hybridisation in situ (ISH): A probe recognizes a particular DNA or RNA sequence, which allows the localization or quantification of this sequence at the cellular level. The cDNA probe is labeled with an isotope or a fluorochrome (FISH)
Quantitative histological analyses
The quantitative information that is extracted from a tissue can be of different types:
- Description of tissue morphology and organization
- Description of cellular organization within a tissue
- Localization and expression of biomarkers in a tissue of in cells
The gain of automated tissue analysis
Automated tissue analysis facilitates:
- Analysis of a unlimited number of histological sections
- Overall quantification of a slide
- Quantification of each tissue structure
- Precise quantification of each cell
- The selection of unusual structures of interest
- Easy navigation through the mass of images
3D Histology
To obtain 3D information in tissue analysis, sequential slides are cut and acquired to perform:
- 3D reconstruction of the object, organ, tissue structure (bio-materials, tumor, vessels)
- Measure 3D volumes
- 3D visualization
3D histology is achieved using image registration tool boxes like ITK or Elastix.
Interested in softwares or services?
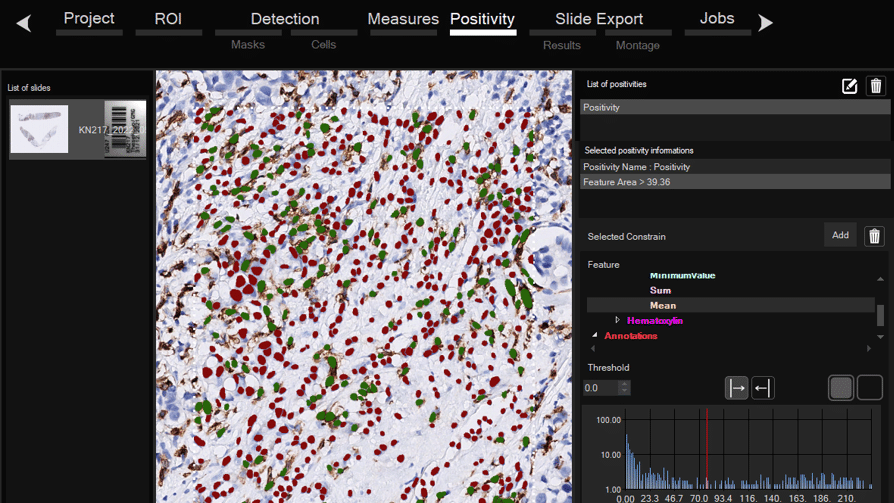
Software for histological samples and TMA
Automatically detects and quantifies your histological slides easily, without requiring bioinformatics expertise. IHC/Fluorescence/Deep learning
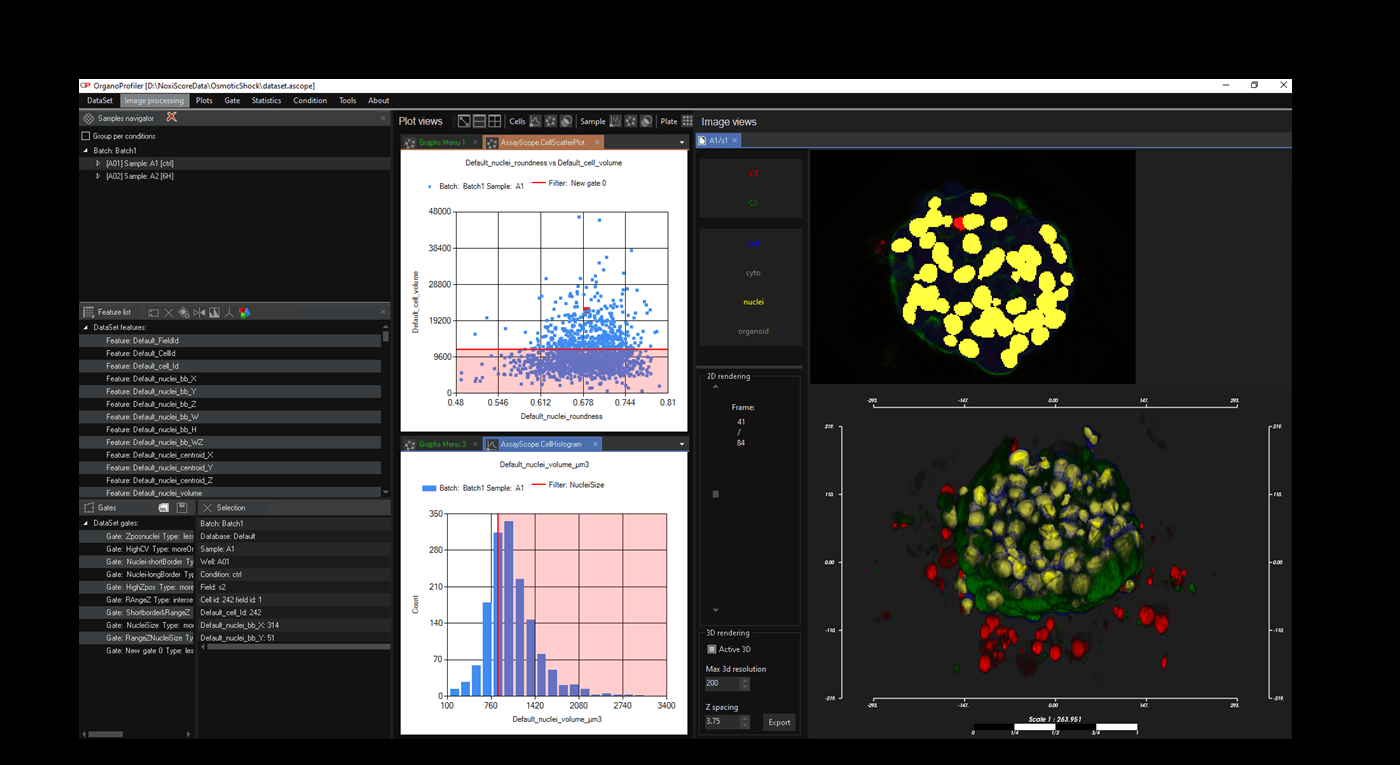
Software for organoid, spheroid and 3D cell culture and tissue
Automatic 3D segmentation and high-content screening analysis software to navigate your assays and monitor drug effects
Services of analysis and characterization
- Image analysis in patients studies
- Characterization of drug/compounds effects
- Automatic detection of rare events

Software development for biomedical imaging
- Customized software solutions with ergonomics UIX
- AI for automatic analysis
- Industrialization of analyses in laboratories
QuantaCell, Campus Balard, IHU
Contact
+33 (0) 9 83 33 81 90
300 av. du Pr Emile Jeanbrau
34090 Montpellier, France